Analysis of multi-condition single-cell data with latent embedding multivariate regression
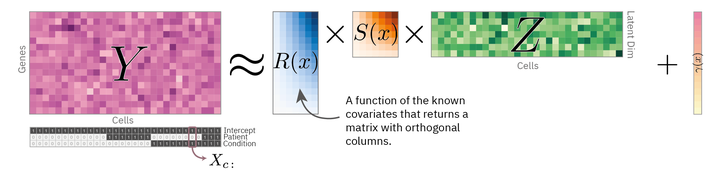 Covariate-dependend matrix factorization
Covariate-dependend matrix factorization
Abstract
Multi-condition single-cell data reveal expression differences between corresponding cell subpopulations in different conditions. Current approaches divide cells into discrete groups or clusters and identify differentially expressed genes between corresponding groups. Here, we propose a method that operates without such grouping. Latent embedding multivariate regression (LEMUR) is based on a parametric mapping of latent space representations into each other and uses a design matrix to encode categorical and continuous covariates. We use the method to analyze a drug treatment experiment on brain tumor biopsies. We detect drug-induced gene expression responses affecting subsets of cells in a continuous latent space representation that does not require discrete categorization of the cells. Latent embedding multivariate regression is a versatile new approach for identifying differentially expressed genes from single-cell data of heterogeneous cell subpopulations or tissues under arbitrary experimental or study designs.