Once upon a time in the world of RNA-seq analysis, there were never enough replicates. Elaborate methods were invented to make the most out of the few that were available (for example: limma, edgeR, DESeq).
Nowadays in times of single cell experiments, the problems are very different. A single experiment can contain 1,000s of cells, but with really shallow sequencing depth. A few years ago there was some discussion if the large number of exact zeros needed to be treated differently. However, recent experiments with negative control data show that there is no zero inflation.
This means the “classical” RNA-seq differential expression tools should still be useful. And indeed, performance benchmarks demonstrate that they perform as good or better than tools specifically developed for single cell data (see for example this paper by Charlotte Soneson and Mark Robinson).
I have recently started my PhD in the group of Wolfgang Huber whose group has developed DESeq and its successor DESeq2. In the benchmark by Soneson and Robinson, it was noticed that DESeq2 scaled badly with an increasing number of cells (Suppl. Figure 26b). To be precise the runtime scaled quadratically with the number of cells, which prohibited DESeq2’s application to large datasets. Thus, my first job as a PhD was to find out why and what could be done about it.
In an earlier blog post, I have described the tools that I used to find the performance bottlenecks. Fortunately, it turned out that the problem was straight forward to fix: in the code there were some diagonal weight matrices that were actually realized although they mostly contained zeros. The solution was to re-arrange some matrix multiplications, so that the whole matrix was never actually needed. All the details can be found in my pull requests to the DESeq2 repository (#PR13 and #PR14).
Mike Love -the maintainer of DESeq2- has already merged my changes into the main repository. DESeq2 now scales linearly with the number of samples. This can mean a huge performance increase if you run DESeq2 with many replicates.
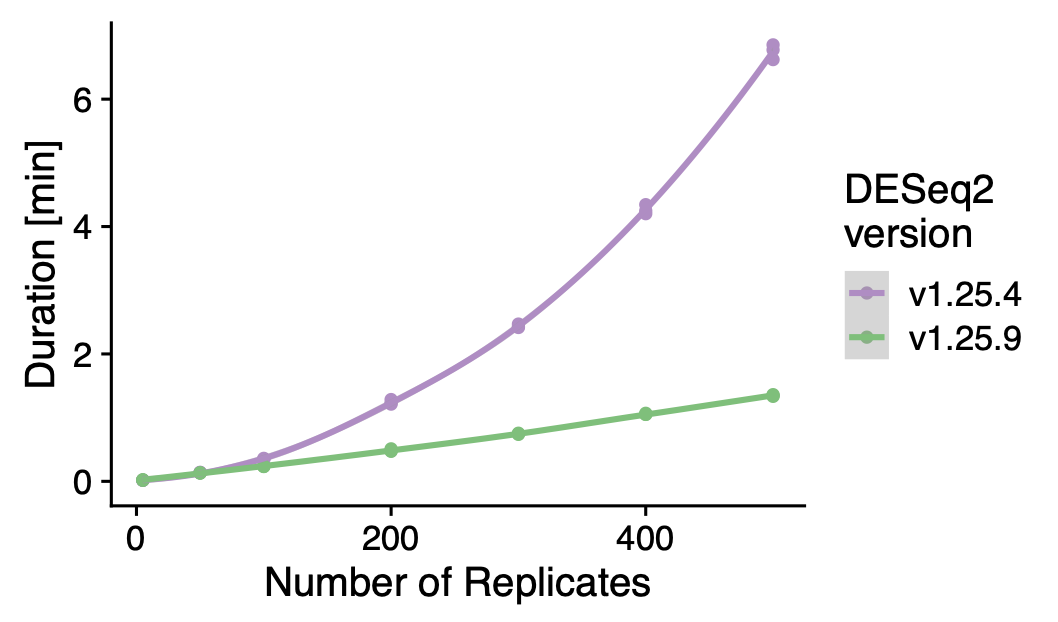
Performance comparison of DESeq2 before and after the performance optimization. For fitting a model with 500 cells, the new version provides a 5x performance boost.
Approximately in October, the new BioConductor version 3.10 will be released and that will include the improved version of DESeq2 . If you cannot wait so long and want to try out the new version today, you can also install the development version from GitHub
devtools::install_github("mikelove/DESeq2")
Changelog:
An earlier version of this blog-post recommended to install an now-outdated version of DESeq2. I removed the specific version specification and recommend to install the latest version of master.
