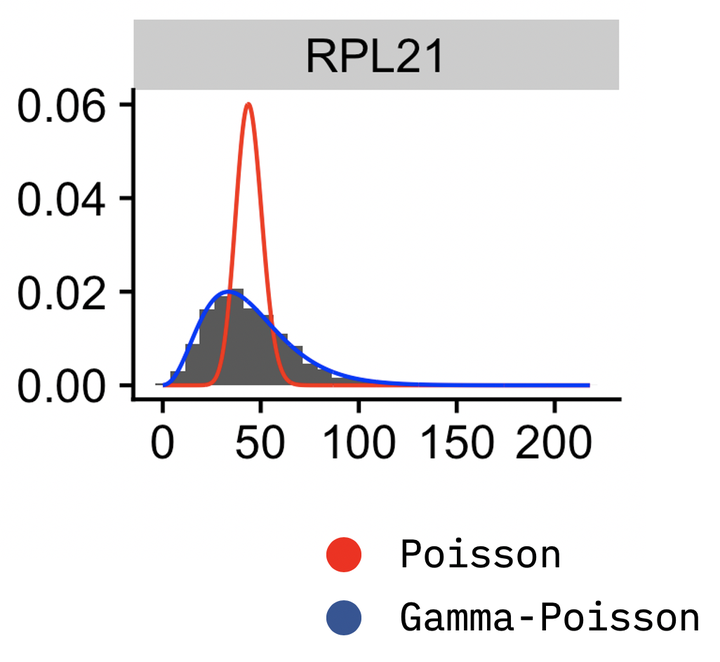
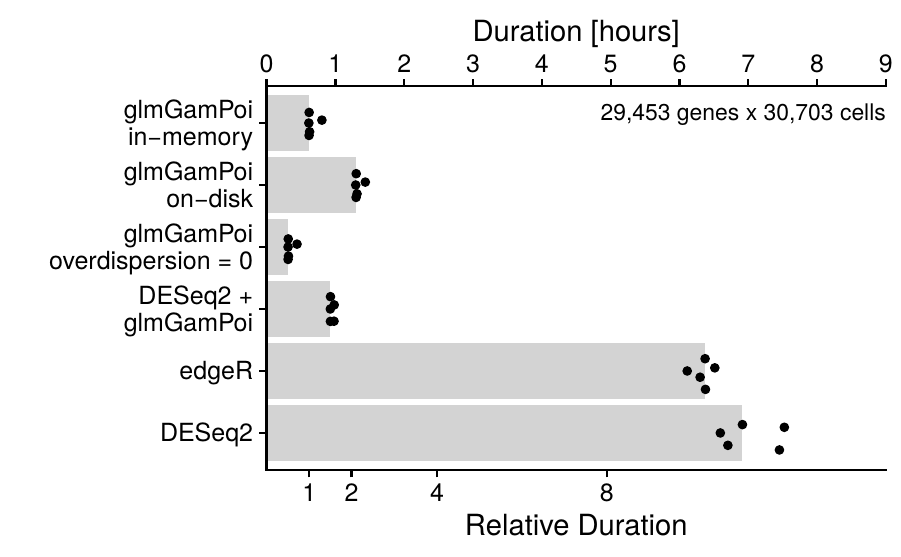
The glmGamPoi package is optimized to fit the Gamma-Poisson^[Gamma-Poisson is an alternative name to Negative Binomial on large single cell datasets. It supports on-disk data via the HDF5Array package and is faster on large datasets than edgeR or DESeq2.
For more information see its Bioconductor page or its Github repository.